Meta-Analytic Research Domains🔗
Article 1 (“Supersizing Meta-Analysis of Psychological Treatment”) offers a basic introduction to the Metapsy “Meta-Analytic Research Domain” (MARD), and presents the results of a comprehensive meta-analysis across world regions and twelve mental disorders. In this section, I provide additional details on the technical infrastructure developed for this initiative, which extends beyond the databases and technical tools mentioned in the article.
MARDs are based on the understanding that a higher level of meta-analytic integration is needed to systematize the surge of randomized controlled evidence in mental health research, and to draw actionable conclusions from it. Formally, MARDs can be described as living systematic reviews of an entire research domain. Unlike conventional systematic reviews and meta-analyses, MARDs do not target one specific population, intervention, comparison, or outcome (PICO). Instead, they cover an entire research field that is too broad for a single meta-analysis (Cuijpers, Miguel, et al., 2022).
MARDs aim to combine the unique strengths of different research synthesis methods. Similar to living systematic reviews (Elliott et al., 2017), they are regularly updated to ensure the most current evidence is available. However, MARDs have a much broader scope than a single “living” meta-analysis or systematic review. Like umbrella reviews (Fusar-Poli & Radua, 2018), MARDs encompass entire research fields while using transparent and consistent data standards. This standardization makes the data easily reusable by the research community and other stakeholders, and it greatly facilitates the development of statistical software and graphical user interfaces (GUIs). These tools are provided as part of a MARD to facilitate rapid evidence generation for an entire research field.
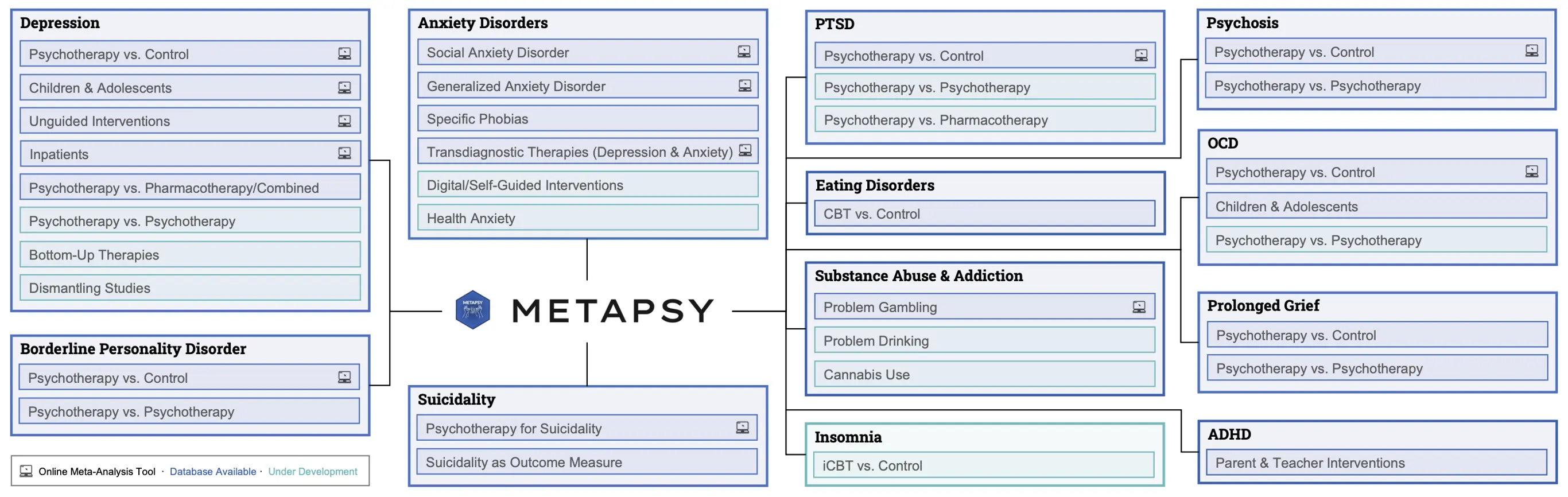
A concrete example of a MARD is the Metapsy initiative (metapsy.org). Metapsy maintains harmonized living databases of psychotherapy trials across all major indications, including depression, anxiety disorders, PTSD, obsessive-compulsive disorder (OCD), borderline personality disorder (BPD), eating disorders, psychosis, problem gambling, prolonged grief, and suicide prevention. Living databases currently under development focus on attention deficit hyperactivity disorder (ADHD), insomnia and health anxiety, among others (see Figure 7). The initiative is based on an international collaboration of research universities and government agencies, including the National Center for PTSD in the United States Department of Veterans Affairs, and the Dutch organization for suicide prevention. The project is coordinated by VU University Amsterdam, and is embedded in the WHO Collaborating Centre for Research and Dissemination of Psychological Interventions. Its data and software infrastructure has been developed at TUM under my lead, and I remain responsible for its technical maintenance.
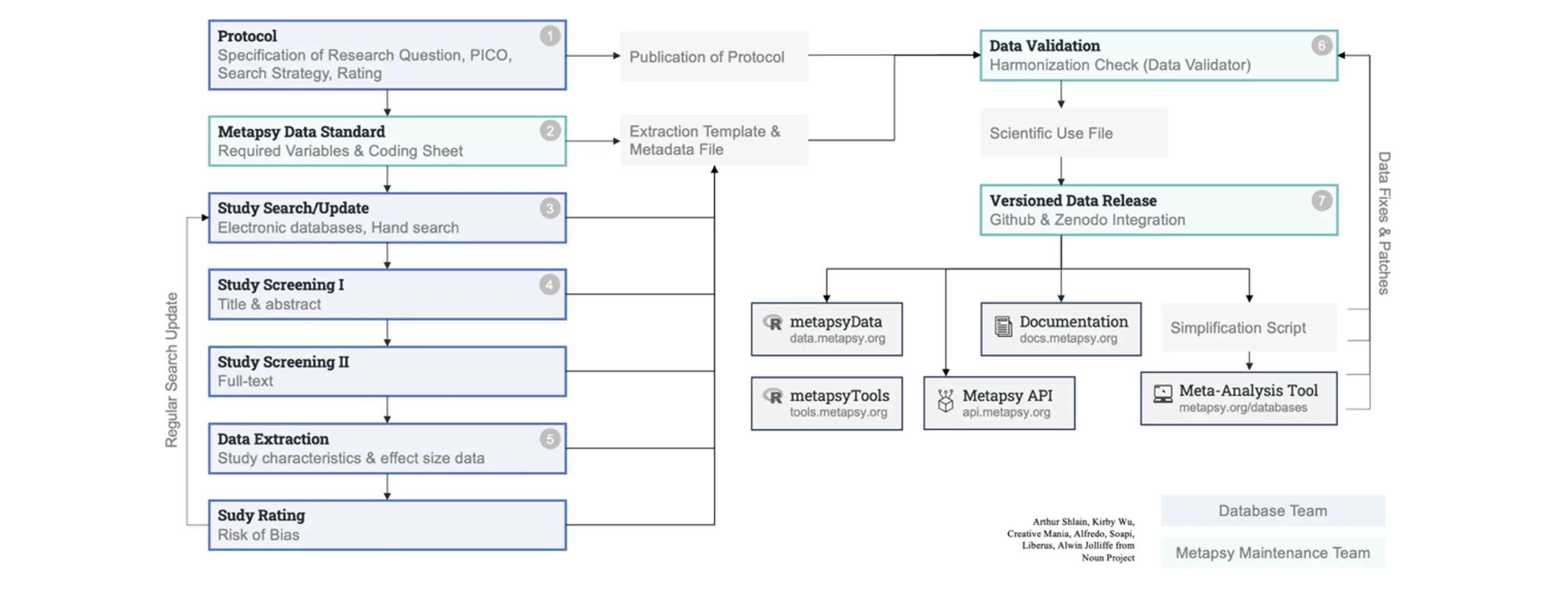
The data release flow and software components of the Metapsy initiative are visualized in Figure 8 above. In the following section, I describe the technical innovations implemented for this project in further detail. These and other descriptions can also be found as supplementary material in Harrer et al. (2024; S4-S6, S10). Details on the searches, data extraction, and coding for each database (Steps 1 and 3-5 in Figure 8 above) are omitted here, but are described in greater detail in Article 1 itself, as well as its supplement (S6).
Data Harmonization & Validation🔗
As part of the Metapsy infrastructure, we have established a uniform formatting standard for all meta-analytic databases. This standard encompasses both the datasets and their metadata, ensuring consistent organization and storage (see Step 2 in Figure 8). The “Metapsy data standard” aligns with the FAIR principles, which stipulate that open data should be findable, accessible, interoperable, and reusable (Wilkinson et al., 2016).
All data collected as part of the Metapsy initiative is compiled in machine-readable formats, such as comma-separated values (CSV) or JavaScript Object Notation (JSON) files (Severance, 2012). This ensures interoperability with other Metapsy software components (e.g., documentation, online tools, R packages, or Application Programming Interfaces) and with external software and programming languages. In each scientific use file (SUF)2Within the Metapsy infrastructure, the term “Scientific Use File” (SUFs; Gollwitzer et al., 2021) refers to subsets of existing databases that are bundled together for public release. SUFs differ from “internal” database versions because they typically have a narrower scope (e.g., focusing on a specific comparator, population, or intervention type). Larger databases are often segmented into several SUFs, each with its own versioned release flow and distinct documentation entry., the core dataset itself is stored in a tabular “wide” format. This means each row contains the effect size data of (i) one comparison in a trial at (ii) a specific time-point, based on (iii) a specific instrument (e.g., the effect of CBT versus waitlist at post-test, using the BDI-II questionnaire, in some trial t); while columns encode the different variables. If a trial only reported one relevant outcome using one instrument at a single assessment point, it will only occupy one row in the data. An exception are multi-arm trials, which contain more than one comparison. In a multi-arm trial with
The Metapsy data standard also encompasses a set of standard variables to be defined in each dataset. This includes extracted study characteristics, as well as a set of calculated effect sizes (Hedges’ g and its standard error; log-transformed risk ratios, their standard error, as well as raw event counts if available). Additionally, the first author’s name, publication year, comparators, assessment point, and instrument are used to create a unique identifier for each row. All variables defined in the Metapsy data standard have fixed and concise names that follow the “tidyverse” style guide (Wickham, 2023). There are also guidelines on other formatting issues such as special characters, leading and trailing whitespaces, or the representation of floats (i.e., decimal numbers), all of which are defined on a documentation webpage (docs.metapsy.org). Collectively, these rules ensure that all datasets are reusable by any kind of software, and can easily be combined with each other.
Lastly, there are specific rules regarding the required metadata. This metadata includes the authors of the database along with their ORCiD identifiers (Haak et al., 2012), the full search strings and date of the last search, the number of included studies and their references, a persistent link to the pre-registration, a description of all included variables, and information needed to populate a Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA; Page et al., 2021) type flow diagram. Each dataset is also assigned a shorthand (e.g., “depression-psyctr”), which serves as a unique identifier. All metadata is stored in a machine-readable JSON or text (TXT) format.
Several tools have been developed to facilitate the correct formatting of datasets. The preparation module of the “metapsyTools” R package (to be covered later) contains functions to check if a dataset conforms to the required data standard, and to convert variables if necessary. Once this step is completed, the package can automatically calculate the effect sizes for each comparison. The functionality of this module has also been integrated into the “Metapsy Data Validator” (tools.metapsy.org/data-validator; see Step 6 in Figure 8). This online tool allows users to (i) upload a dataset, (ii) check if it adheres to the data standard, (iii) resolve any remaining conflicts, and then (iv) calculates the effect sizes automatically. In the end, users can download a formatted version of the data that is ready to be released on the Metapsy platform.
Data Release & Versioning🔗
Within the Metapsy infrastructure, all datasets are released through a workflow based on the version control system Git (Blischak et al., 2016; see Step 7 in Figure 8). This system allows to preserve every stage of a database over time, ensuring that previous versions are never lost and can be easily reproduced. Git is a standard tool in software development and is also recommended for open science (Millman & Pérez, 2014). Although its infrastructure is not optimized for large file storage (e.g., > 2 gigabytes), this is not an issue for meta-analytic data, where even large databases hardly exceed 10 megabytes. Using Git, all datasets and their metadata are uploaded to an online repository hosted by the Metapsy project organization (github.com/metapsy-project).
Once a dataset is ready to be integrated, the current status of the repository is turned into a release: a persistent version of the database that others can directly access. During this process, a dataset is assigned a version number that follows the major-minor-patch structure of the Semantic Versioning 2.0.0 standard (semver.org/spec/v2.0.0), yet with a few adaptations. The major number indicates the last two digits of the year in which a version of the dataset was released (e.g., “22” if the release was in 2022). The minor number is reserved for changes and updates that may impact the results users obtain when analyzing the data. This includes changes in the calculated effect sizes or the inclusion of new studies. The patch number is used for minor fixes that will not impact the results, such as minor typo corrections.
Therefore, the first version of a dataset released in 2024 is numbered 24.0.0, with 24.0.1 for a subsequent patch, and 24.1.0 for a minor change. Using years for versioning is not typical, but this approach is used in the Metapsy initiative because most databases are updated yearly. This allows for a quick association of version numbers with the year of their release.
New releases created by the Metapsy organization are automatically indexed on Zenodo (zenodo.org), an all-purpose open repository operated by the European Organization for Nuclear Research (CERN). At this step, the dataset version is assigned a digital object identifier (DOI), and a record of the dataset and its metadata is stored on the CERN servers. This means that the datasets and metadata are openly available for everyone, and can be easily retrieved using their unique identifier. Zenodo not only creates a unique DOI for a specific dataset version but also a “concept DOI” that can be used to reference a dataset in general, and which always defaults to the latest version.
Zenodo also maintains its own Application Programming Interface (API; developers.zenodo.org). This allows all software in Metapsy to automatically incorporate new datasets and their metadata within seconds after their release, which is particularly helpful for maintaining an up-to-date documentation of each database online docs.metapsy.org.
Statistical Software & APIs🔗
Two statistical software packages have been developed for the Metapsy infrastructure: metapsyData (Harrer, Sprenger, et al., 2022) and metapsyTools (Harrer, Kuper, et al., 2022). Both of these packages are implemented for the statistical programming language R, and are openly accessible as Free/Libre Open-Source Software under an MIT license (FLOSS; Fortunato & Galassi, 2021). While Article 1 provides a brief description of metapsyData and metapsyTools, this section offers more detailed technical information on their functionality.
The metapsyData package is designed to provide a lightweight interface for researchers to import versioned database releases into their R environment. It includes two main functions: “listData” and “getData”. The “listData” function prints an overview of all available datasets and their shorthand identifiers. These shorthands can then be used in the “getData” function to retrieve specific datasets and their metadata. The version argument in “getData” allows users to retrieve specific versions of the database, which is recommended to ensure that R scripts remain fully reproducible, even when new updates are released. The resulting data object follows an object-oriented programming paradigm, implementing the R6 reference class (Chang, 2022). This means the object returned by “getData” comes pre-packaged with the downloaded dataset, metadata, and some helper functions. For instance, the “returnMetadata” function collects all the metadata stored in the object and returns it as a list within the R environment, while the “variableDescription” function provides a named list object describing each included variable. A designated documentation page has been created for the package, which can be found at data.metapsy.org.
The metapsyTools package offers a comprehensive set of tools to (i) compile a database according to the Metapsy data standard (“preparation module”) and (ii) analyze databases following the Metapsy data standard, using state-of-the-art meta-analytic methods (“analysis module”). The high level of standardization provided by the Metapsy data standard enables the package to automate many parts of a typical meta-analysis pipeline. The “analysis module” adopts a tidyverse-style coding approach, where each step of a meta-analysis is represented by a distinct function, and different analysis steps are connected using pipe operators (Wickham et al., 2023). Initially, the “filterPoolingData” and “filterPriorityRule” functions can be used to filter relevant comparisons from a database using logical operators, fuzzy string matching, or prioritization rules. The “runMetaAnalysis” wrapper function then automatically executes conventional and advanced meta-analytic models using the data. Several “S3 methods” are defined for this function, enabling the inclusion of fixed predictors in a meta-regression, conducting subgroup analyses, or generating forest plots. Once the models are fitted, the effects can also be examined while controlling for small-study effects and potential publication bias, using the “correctPublicationBias” function.
The metapsyTools package imports core functionalities from the meta (Balduzzi et al., 2019) and metafor (Viechtbauer, 2010) packages. In particular, the “metagen,” “rma.uni,” and “rma.mv” functions are called internally to fit most of the models provided in metapsyTools. This also facilitates the integration of new functionalities for these packages (both of which are actively maintained) into metapsyTools. The “runMetaAnalysis” function saves all fitted models internally within a returned object. These internal objects function like regular “meta” or “metafor” models, which allows the use of any functionalities developed for these packages, even if not directly implemented in metapsyTools. Other relevant imports come from the dmetar (Harrer et al., 2019), metasens (Schwarzer et al., 2022), and clubSandwich (Pustejovsky, 2022) packages.
The metapsyTools package includes a designated documentation webpage, available at tools.metapsy.org (see Figure 9). This documentation page also provides an overview of all meta-analytic models and sensitivity analyses integrated into the package (see tools.metapsy.org/articles/web/get-started#pooling-effects). By default, all these models are run simultaneously, and a summary of the results is presented. This allows users to assess the robustness of the effect across more basic and complex multivariate meta-analytic models, commonly used sensitivity analyses, and methods to correct for publication bias. Using the “subgroupAnalysis” and “metaRegression” methods, it is also possible to examine predictors of differential treatment effects. All summaries in metapsyTools are returned as pre-formatted HTML tables, which can be easily copied and pasted into a word processor (e.g., MS Word), facilitating the reporting of results. Similarly, the “createStudyTable” function creates an overview table of the included studies, as required by the PRISMA guidelines (item 17) and APA Meta-Analysis Reporting Standards (MARS; APA, 2008).
The metapsyData and metapsyTools packages can only be used within an R environment. For developers who wish to utilize the databases in other programming languages or environments, the Metapsy initiative also offers its own Application Programming Interface (API; metapsy.org/api). Unlike graphical user interfaces, which facilitate human-computer interaction, APIs enable communication between two different software systems, typically over the Internet. APIs are essential components of nearly all modern web applications and are increasingly used for research purposes (Lelong et al., 2022; Swaminathan et al., 2016). The Metapsy API provides two endpoints: one to list all available datasets, and another to retrieve specific dataset versions. All response bodies returned by the API are JSON-encoded. Through the API, Metapsy data can be imported in a machine-readable format from any programming language, facilitating the development of new software based on the databases. This software component was implemented to enhance the interoperability of the Metapsy infrastructure with other applications and workflows.

Online Meta-Analysis Tools🔗
The Metapsy initiative also offers a comprehensive suite of user-friendly graphical interfaces (GUIs). These include an online meta-analysis tool available for all released databases, accessible through the project homepage (metapsy.org/databases; see Figure 10). The tools operate on the most recent version of each dataset, allowing users to further refine the data using buttons and sliders to address specific research questions. For instance, users can filter studies by publication year, focus on specific interventions or population groups, or limit results to studies with a low risk of bias.
The tool then automatically applies various meta-analytic methods to the selected data, and graphically presents the results. Users can generate a PDF report summarizing the findings, with the analyzed data appended and assigned a unique tracking ID. All applied filters are documented to ensure transparency and reproducibility, helping others confirm that the findings are not the result of selective data manipulation. The report also includes references for all included studies, as well as a technical summary of the R and package versions used.
The Metapsy homepage also offers a meta-analytic “effect explorer” (metapsy.org/database/explore; Cuijpers, Miguel, Ciharova, Harrer, et al., 2023; see Figure 10) designed for patients and clinicians. This tool allows users to examine response rates of various psychological treatments for eight mental disorders, providing results in lay terms, and with a minimalistic design. To support these online tools, Metapsy maintains a dedicated Linux server (metapsy.dev), currently running Ubuntu 22.04 x64, R (version 4.2.2), Python (version 3.10.12), shiny-server (Posit, 2023), and TeX Live (TeX Users Group, 2023). This setup ensures easy deployment of new tools, as well as scalability of existing applications as user demand grows.
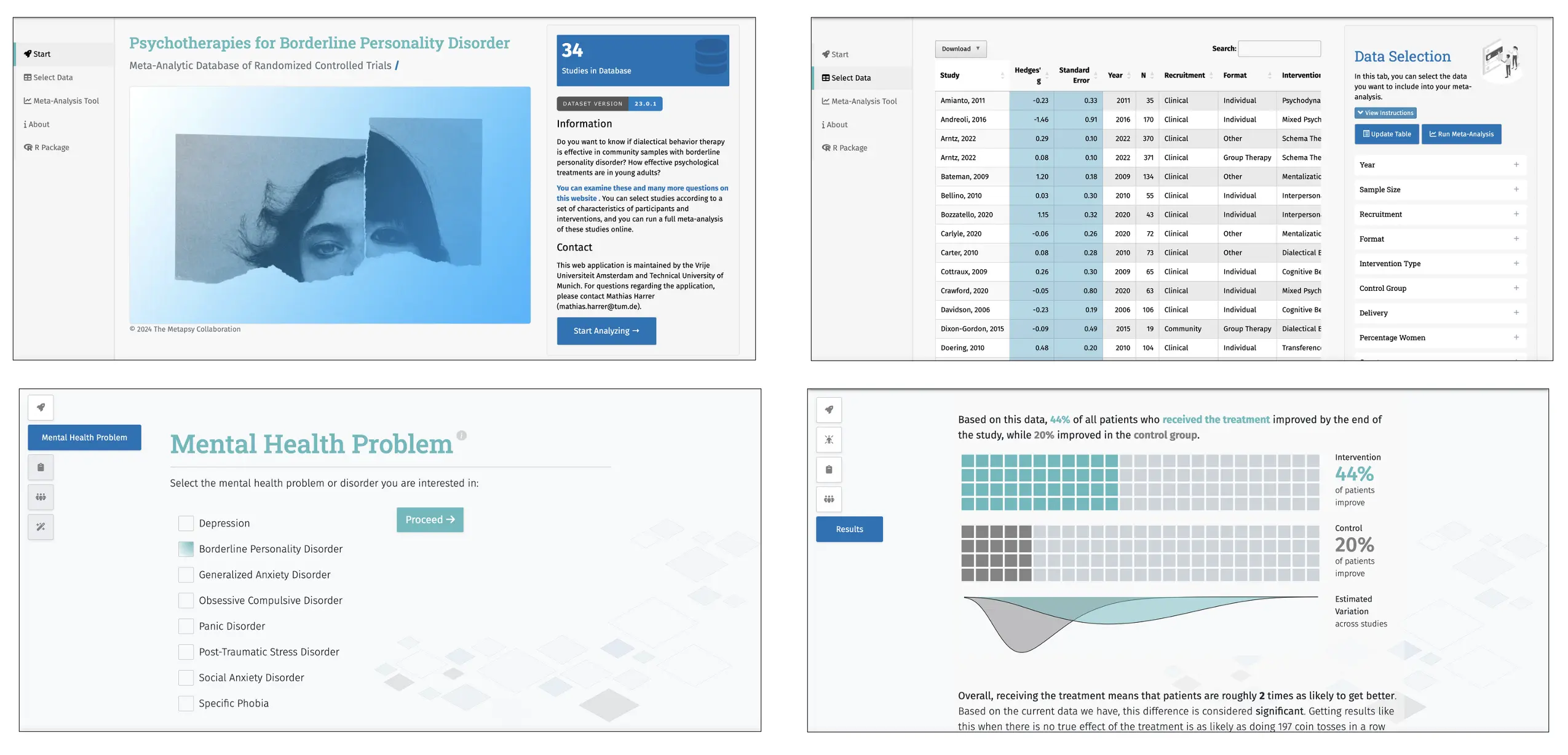
The GUIs developed for the Metapsy MARD aim to assist a wide range of health professionals; including guideline developers. When creating or updating treatment guidelines, it is crucial to identify the current evidence base of interventions for specific patient groups. For example, when developing guidelines for older patients with subthreshold depression, guideline developers may often rely on meta-analytic evidence to recommend treatments. Yet, existing reviews may not fully address this specific research question, may be outdated, or a relevant meta-analysis might not exist. The online meta-analysis tools provided by Metapsy could help overcome these challenges. Guideline developers could use the online depression psychotherapy tool (metapsy.org/database/depression-psychotherapy) to filter relevant studies, and generate a meta-analysis comparing different treatments. The report generator can then be used to document the findings. Additionally, the general public could benefit from tools like the Metapsy “effect explorer” to learn more about the efficacy of psychological treatments for various disorders, and to better understand the robustness of our current knowledge.