Meta-Analytic Predictive Modeling🔗
In Articles 3 (“Meta-Analytic Predictive Modeling”) and 4 (“Meta-Analytic Decision-Tree Learning”), I employ new types of multivariable prediction models to establish “precision treatment rules” (PTRs) for two digital psychological interventions. These models are developed in a “meta-analytic” context, combining parametric multi-level modeling with flexible machine learning algorithms to provide personalized predictions. This approach uses data from multiple studies, thus providing a form of “data fusion” as described by Bareinboim & Pearl (2016). The present section gives a detailed technical description of the developed models, their validation, and the strengths of the meta-analytic approach; including its ability to evaluate performance across various settings and contexts. These and further descriptions have also been added as supplementary material in Harrer et al. (2023).
In Article 3, precision treatment rules (PTRs) are developed for a digital stress intervention when used as an indirect treatment for depression. Individual participant data (IPD) from six randomized controlled trials were available in the development sample, and the target outcome was the effect on depressive symptom severity after six months. Differential treatment benefits were predicted based on 20 self-reported indicators assessed at baseline across all six trials, with sporadically missing values handled using multiple imputation. Indicators were selected a priori on theoretical grounds and based on existing scientific evidence (see the methods section in Article 3 for further details).
Using terminology of the Predictive Approaches to Treatment Effect Heterogeneity (PATH) Statement (Kent, Paulus, et al., 2020; Kent, Van Klaveren, et al., 2020), an “effect modeling” approach was used to develop the main prediction model in this study. Prognostic variables were incorporated into a Linear Mixed Model (LMM) with a random trial intercept (
In this model,
Additionally, I explored another model incorporating post-randomization interim assessments. This approach enables updated predictions of long-term outcomes several weeks into treatment, facilitating the initiation of further measures if no significant symptom reduction is expected. Conversely, for patients who did not receive the intervention, such models could help evaluate the need for treatment after a “watchful waiting” period. In all included trials, interim assessments of depressive symptom severity were conducted seven weeks post-randomization. These measurements were included as an additional prognostic and prescriptive predictor to create the “update” model (
As part of the effect modeling approach described earlier, multiple interaction terms are incorporated into the model equation. This renders the model susceptible to common issues found in conventional subgroup analyses, such as low power and micronumerosity (Brookes et al., 2001; Dahabreh et al., 2016). Consequently, as a sensitivity analysis, a risk modeling approach was also employed, using all baseline variables as predictors (Kent, Paulus, et al., 2020). In this model, a linear predictor of risk (
Three methods were used to estimate parameter values and compare their individual performance for each model equation. First, a “full model” linear mixed model (LMM) was fitted using penalized quasi-likelihood (PQL), as implemented in the “glmmPQL” function of the R package MASS (Venables & Ripley, 2002; Wolfinger & O’Connell, 1993). Second, LMMs were maximized using
Third, each model was estimated using the novel likelihood-based boosting for linear mixed models (
To internally validate the models, I employed methods to assess their (i) apparent performance, (ii) optimism-adjusted performance, and (iii) transportability. For each model, the apparent performance was determined by calculating the coefficient of determination (
To derive an optimism-adjusted estimate of each model’s predictive performance, I applied an adapted version of the bootstrap bias correction procedure for IPD meta-analytic settings (Riley et al., 2021; chap. 7.1.7). A total of
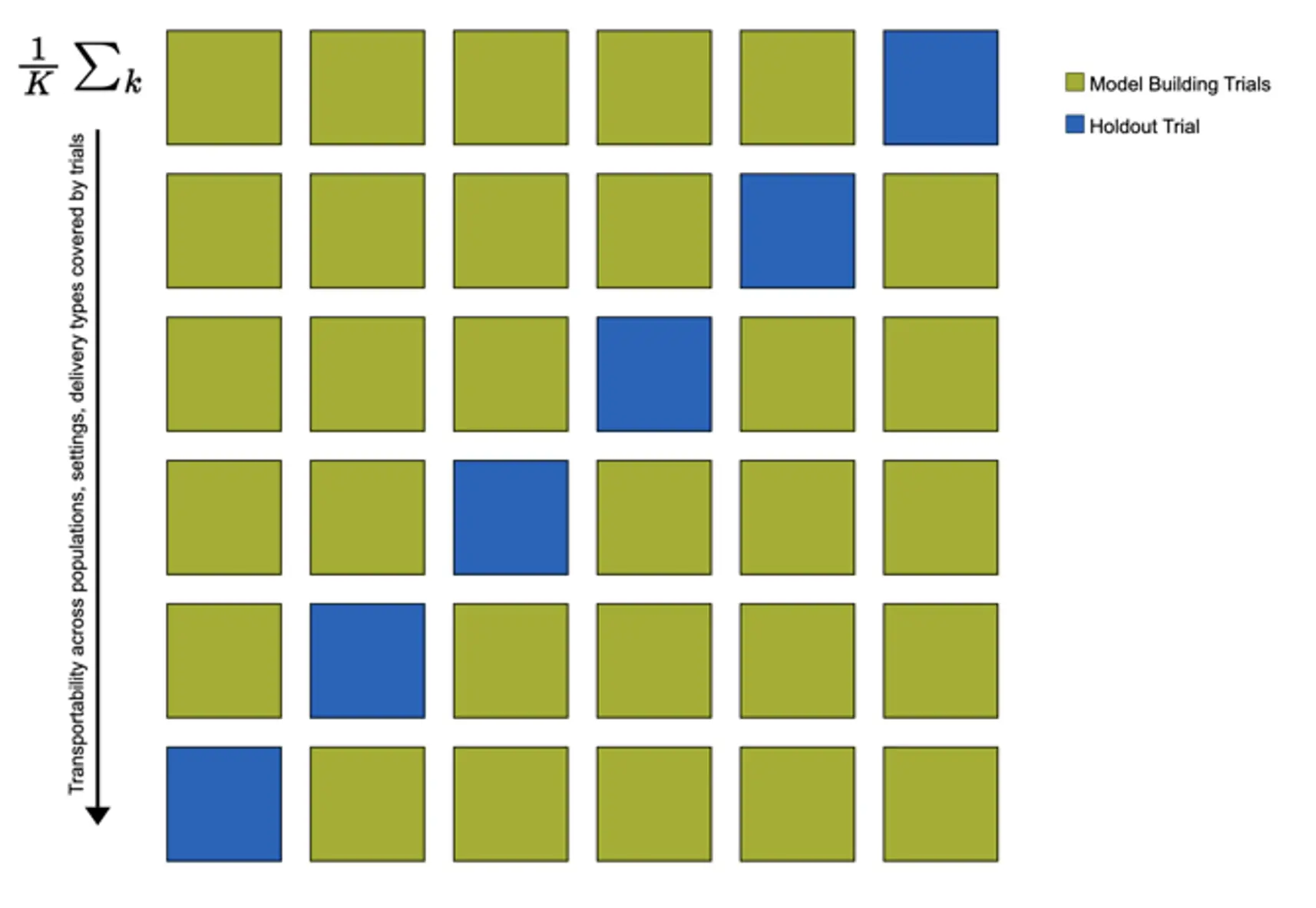
In the study, there was some variation in the studied population (case-mix), setting, and implementation of the digital stress intervention across trials. This allowed me to examine the transportability of the developed models, i.e., the potential generalizability of model predictions in populations external to those used for model development (Riley et al., 2021). I used “internal-external” cross-validation to estimate each model’s transportability. This means that models' predictive performance is estimated in
It is frequently stressed that predictive accuracy alone is not informative about the clinical utility of a prognostic model (Steyerberg, 2019; Kent, Van Klaveren, et al., 2020; Vickers et al., 2007). Predictions of predictive models should allow for improved clinical decision-making, i.e., the discrimination between future patients who do or do not benefit from the intervention. Thus, I additionally conducted a clinical usefulness analysis focusing on conditional average treatment effects (CATE; Li et al., 2018), meaning benefit scores predicted by the main models
The approach I employed heavily draws on the Neyman-Rubin Causal Model (NRCM) (Aronow & Miller, 2019). Adopting Rubin’s (1974) potential outcome framework, CATEs can be viewed as estimates of the expected causal effect
In our case, they may be more elegantly expressed as individualized estimates of the (standardized) between-group intervention effect size,
We can also derive the overall benefit
It is possible to calculate empirical versions of these entities (
Subgroup-conditional effects were estimated based on two clinically relevant, a priori determined thresholds. First, I assumed a cut-point of
To be clinically useful, the precision treatment rule should assign patients with clinically relevant treatment benefits to the “treatment recommended” subgroup, while patients with clinically negligible or no treatment benefits are allocated to the “treatment not recommended” subgroup. Thus, upon implementation of the treatment assignment rule, the bootstrap-bias corrected estimate of the treatment effect/benefit in the “treatment not recommended” subgroup should be clinically negligible (i.e., too small to warrant assigning the intervention from a clinical standpoint), while the estimated effect in the “treatment recommended” group should be high.
I assumed that nearly all CATEs are positive; i.e., that patients for whom the intervention is genuinely harmful compared to no treatment are very rare or nonexistent. Therefore, for the treatment rule to be useful, estimates of its overall benefit
In Article 4, algorithmic modeling was used to examine heterogeneous treatment effects of a digital intervention for depression in back pain patients. The target outcome was effects on PHQ-9 depressive symptom severity at 9-week post-test. Two RCTs were available for this study, one focusing on subthreshold depression patients, and the other on patients with a clinical diagnosis of MDD (Baumeister et al., 2021; Sander et al., 2020). The same intervention was applied in both trials. In our secondary analysis, we employed decision tree learning to detect subgroups with differential treatment effects. Our model was externally validated in a third trial among back pain patients with depression who are currently on sick leave (Schlicker et al., 2020). As before, we combined algorithms derived from machine learning with parametric multi-level models, employing “multi-level model-based recursive partitioning” (MOB; Fokkema et al., 2018). Parts of the explanations below can also be found in Harrer, Ebert, Kuper et al. (2023).
In this study, a data-driven approach was used to select relevant baseline indicators for further modeling steps. Putative moderators were entered into the random forest methodology for model-based recursive partitioning (
Multi-level model-based recursive partitioning was then used to obtain a final decision tree predicting differential treatment effects. MOB can be applied to any parametric model fitted using
In our study, the stopping rule of this process was defined by setting the significance level for parameter stability tests to
A computational challenge in this study was that only two trials were available in the development set, which means that estimation of between-study heterogeneity variances
Where
This adapted model was then used in the nodes while employing the recursive portioning routine.